The American Journal of Human Genetics (AJHG) has awarded Yosuke Tanigawa, research scientist and former postdoc at MIT CSAIL and the Broad Institute of MIT and Harvard, the AJHG Award for Outstanding Trainee Publication. Tanigawa received the honor for leading the development of a genetic prediction model that accounts for a wider diversity of genetic ancestries worldwide. He was recognized at the award ceremony at the annual meeting of the American Society of Human Genetics (ASHG).
The AJHG Award is given annually to two researchers who’ve contributed to the field of human genetics with an impactful research paper early in their careers. Having started at MIT in 2021, Tanigawa has focused his research on computational biology and statistical models that predict a patient’s risk for conditions like heart disease and physical traits like being tall. He has developed several computational and statistical approaches that dive deeper into disease heterogeneity — differences among individuals in how manifestations of diseases are influenced by different mechanisms — and uses them to analyze large-scale data of patients from different genetic backgrounds.
Tanigawa earned this honor for his work on the “inclusive polygenic score (iPGS),” the first genetic prediction model applicable to everyone regardless of their genetic ancestry. The paper showed the potential to increase the accuracy of polygenic scores—a genetic forecast for cumulative predispositions to medical conditions like diabetes and heart attack, potentially enabling more efficient disease prevention. This model was especially impactful for groups traditionally underrepresented in genetic studies, where iPGS showed a marked improvement across the board.
The research authored by Tanigawa and Manolis Kellis, a professor of computer science at MIT CSAIL, incorporated genetic data from over 280,000 people and another set of roughly 81,000 held-out individuals from the UK Biobank. They found that by incorporating more diverse genetic data, their model was more accurate across each group compared to models trained only on European-ancestry individuals. The team’s iPGS model saw 61% average improvements for African ancestry and 11% for South Asian descent, for example.
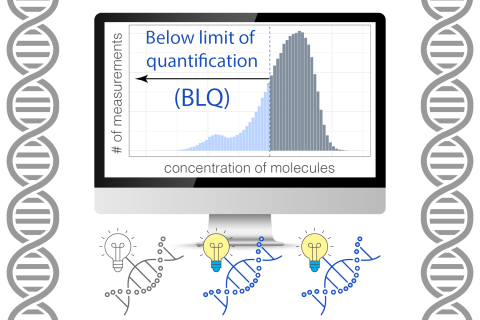
Recently, Tanigawa and his colleagues have advanced several research projects. On the one hand, he has just published a new study in AJHG, investigating typically discarded measurements — data points below the limit of quantification — to enhance genetic discovery. On the other hand, he has been leading several efforts in polygenic score research. For example, he recently co-led another research project: “GenoBoost,” a flexible framework that estimates individuals’ polygenic risk scores by going beyond simply adding up alleles and incorporating nonlinear effects. One of his follow-up studies, which incorporates the insights from GenoBoost into iPGS, has been presented at the annual meeting and selected as a semifinalist for the ASHG Trainee Research Excellence Award. In parallel, he and his colleagues recently developed a new approach to incorporate biological priors to build more reliable iPGS models, which was also presented as an oral platform presentation. Those efforts help expand the beneficiaries of genomics research and realize precision medicine.
Tanigawa’s AJHG Award adds to his honors list since coming to MIT. In 2022, he received ASHG’s Charles J. Epstein Trainee Award for Excellence in Human Genetics Research and MIT Technology Review’s Innovators Under 35 Japan Award. More recently, he received the MIT Prize for Open Data for making iPGS models more accessible.