Much of the research aimed at understanding the root causes of biological differences has focused on differences in the genes. But in recent years there has been an increasing emphasis on studying the changes in the regulatory switches in our DNA that control how genes are turned on and off.
A key technology that enables these studies is profiling of the epigenome, the set of physical and chemical modifications on the DNA and its packaging. The epigenome is particularly important because it is influenced both by the DNA code that we inherit, but also by the environment that we grow up and live in. In addition, changes in our epigenome in different conditions and in different cell types can provide crucial information about how the DNA is used.
“Many computational methods have been developed that compare the activity of genes”, says senior author Manolis Kellis, a professor of computer science at CSAIL and at the Broad Institute of MIT and Harvard. “However computational methods that compare epigenomic activity are still scarce, thus limiting our ability to understand how the epigenome changes across cell types, individuals, or human disease.”
In a paper published this week in the peer-reviewed journal Nature Communications, CSAIL researchers present a new computational method for systematically comparing large numbers of epigenome maps, to recognize the signature characteristics of different types of cellular properties.
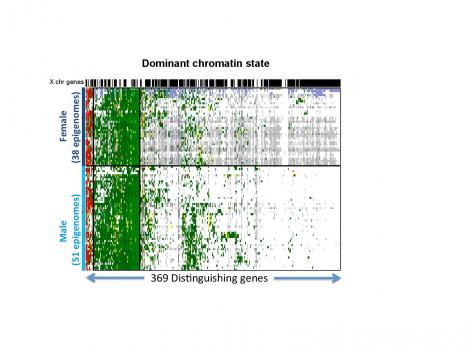
The team developed a new method, ChromDiff, that applies rigorous statistical techniques for comparing groups of epigenomic annotations, and they applied it to the largest collection of reference epigenomes to date, published earlier this year as part of the Roadmap Epigenomics project.
The method found that diverse epigenomic differences distinguish male vs. female cells, young vs. old cells, and blood vs. tissue samples. Some of these differences were also reflected in expression changes, but many were not, emphasizing the richness of epigenomic information. The team says that it could use ChromDiff to identify genes and environmental changes that may play a role in various diseases, such as Alzheimer’s disease, blood cancers like leukemia, and gastric diseases.
“We are optimistic that our method has significant potential for being able to help scientists better understand epigenomic effects, which can be applied to understand epigenomic differences in the onset and progression of cancer or other human diseases,” says first author Angela Yen, a graduate student at CSAIL.
The work comes on the heels of Kellis’ efforts leading a multi-university team to develop a comprehensive map of the human epigenome that has opened up new possibilities for understanding the circuitry of the human genome, and the diversity of human cell types.
The epigenome
The epigenome is extremely important when comparing different types of cells. All the cells in one person’s body, ranging from skin cells to blood cells to muscle cells, have identical DNA, so the environment of the DNA can serve as a “map” that gives us hints about how that DNA was used.
In some cell types, a gene might play an important role, while in other cell types, the same gene might be completely silent, as it is not needed. The machinery that controls the gene activity in this way can be influenced by or leave clues in the DNA environment.
“When biologists study the DNA environment, they can then trace back to what happened with the machinery, in the same way that hunters can study footprints to guess where an animal was and what it did,” says Yen.
The team has put ChromDiff online, to allow other researchers to analyze their own data.
“We hope that our results, as well as results from other scientists who use ChromDiff, will contribute to our larger understanding of the human epigenome and how it relates to the biological properties of different cell and tissue types,” says Yen. “In the big picture, we believe our results and our method have the potential to help guide scientists towards a deeper understanding of gene regulation, cellular biology, and ultimately human disease.”